Gaussian Process Classification Model in various PPLs
This page was last updated on 29 Mar, 2021.
As a follow up to the previous post, this post demonstrates how Gaussian Process (GP) models for binary classification are specified in various probabilistic programming languages (PPLs), including Turing, STAN, tensorflow-probability, Pyro, Numpyro.
Data
We will use the following dataset for this tutorial.
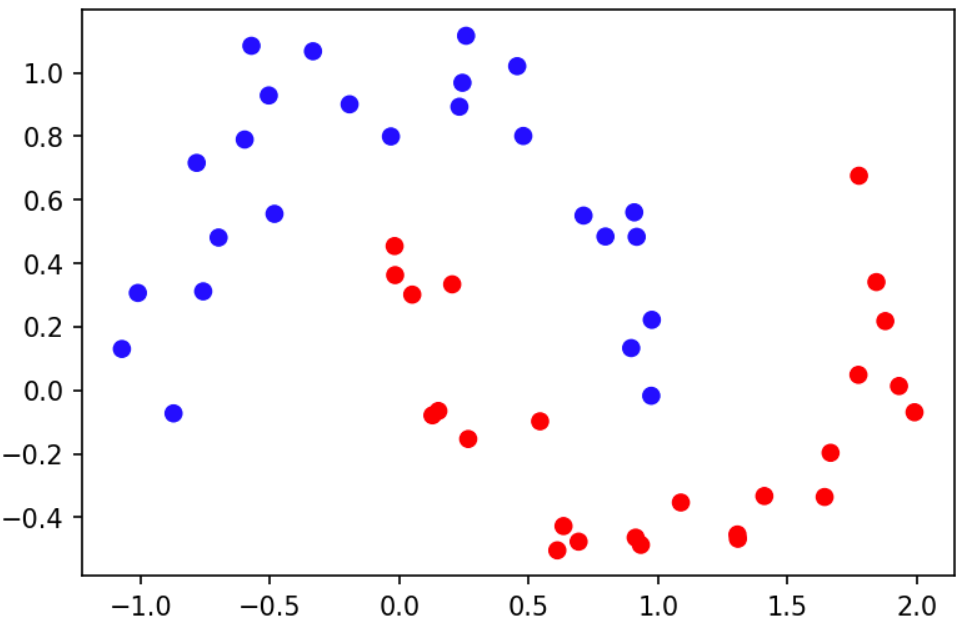
This dataset was generated using make_moons from the sklearn python
library. The input ($X$) is a two-dimensional, and the response ($y$) is
binary (blue=0, red=1). 25 responses are 0, and 25 response are 1. The goal,
given this dataset, is to predict the response at new locations. We will use a
GP binary classifier for this task.
Model
The model is specified as follows:
\[\begin{aligned} y_n \mid p_n &\sim \text{Bernoulli}(p_n), \text{ for } n=1,\dots, N &(1)\\ \text{logit}(\bm{p}) \mid \beta, \alpha, \rho &\sim \text{MvNormal}(\beta \cdot \bm{1}_N, \bm{K}_{\alpha, \rho}) &(2) \\ \beta &\sim \text{Normal(0, 1)} &(3) \\ \alpha &\sim \text{LogNormal}(0, 1) &(4)\\ \rho &\sim \text{LogNormal}(0, 1) &(5) \end{aligned}\]We use a Bernoulli likelihood as the response is binary. We model the logit of the probabilities in the likelihood with a GP, with a $\beta$-mean mean function and squared-exponential covariance function, parameterized by amplitude $\alpha$ and range $\rho$, and with 2-dimensional predictors $x_i$. The finite-dimensional distribution can be expressed as in (2), where $K_{i,j}=\alpha^2 \cdot \exp\bc{-\norm{\bm{x}_i - \bm{x}_j}^2_2/2\rho^2}$. The model specification is completed by placing moderately informative priors on mean and covariance function parameters. For this (differentiable) model, full Bayesian inference can be done using generic inference algorithms (e.g.ADVI, HMC, and NUTS).
None of the PPLs explored currently support inference for full latent GPs with non-Gaussian likelihoods for ADVI/HMC/NUTS. (Though, some PPLs support variational inference via variational inference for sparse GPs, aka predictive processe GPs.) In addition, inference via ADVI/HMC/NUTS using the model specification as written above leads to slow mixing. Below is a reparameterized model which is (equivalent and) much easier to sample from using ADVI/HMC/NUTS. Note the introduction of auxiliary variables $\boldsymbol\eta$ to achieve this purpose.
\[\begin{aligned} y_n \mid p_n &\sim \text{Bernoulli}(p_n), \text{ for } n=1,\dots, N \\ \text{logit}(\bm{p}) &= \bm{L} \cdot \boldsymbol{\eta} + \beta\cdot\bm{1}_N, \text{ where } \bm{L} = \text{cholesky}(\bm{K}) \\ \eta_n &\sim \text{Normal(0, 1)}, \text{ for } n=1,\dots,N \\ \beta &\sim \text{Normal(0, 1)} \\ \alpha &\sim \text{LogNormal}(0, 1) \\ \rho &\sim \text{LogNormal}(0, 1) \end{aligned}\]PPL Comparisons
Below are snippets of how this model is specified in Turing, STAN, TFP, Pyro, and Numpyro. Full examples are included in links above the snippets. The model was fit via ADVI, HMC, and NUTS for each PPL. The inferences were similar across each PPL via HMC and NUTS. The results were slightly different for $\alpha$ using ADVI, but were consistent across all PPLs. Below, we present posterior summaries for NUTS from Turing.
Here are some algorithm settings used for inference:
- ADVI
- Number of ELBO samples was set to 1
- Number of iterations was set to 1000
- Number of samples drawn from variational posterior distribution = 500
- HMC
- Step size = 0.05
- Number of leapfrog steps = 20
- Number of burn-in iterations = 500
- Number of subsequent samples collected = 500
- NUTS
- Maximum tree depth = 10
- Target acceptance rate = 80%
- Adaptation / burn-in period = 500 iterations
- Number of sampler collected = 500
Results
Below, the top left figure is the posterior predictive mean function $\bm{p}$ over a fine location grid. The data is included for reference. Note that where data-response is predominantly 0 (blue), the probability of predicting 0 is high (indicated by low probability of predicting 1 at those locations). Similarly, where data-response is predominantly 1 (red), the probability of predicting 1 is high. In the regions between, the probability of predicting 1 is near 0.5. The top right figure shows that where there is ample data, uncertainty (described via posterior predictive standard deviation) is low (whiter hue); and where data is lacking, uncertainty is high (darker hue). The bottom three panels show the posterior distribution of the GP parameters, $(\rho, \alpha, \beta)$.
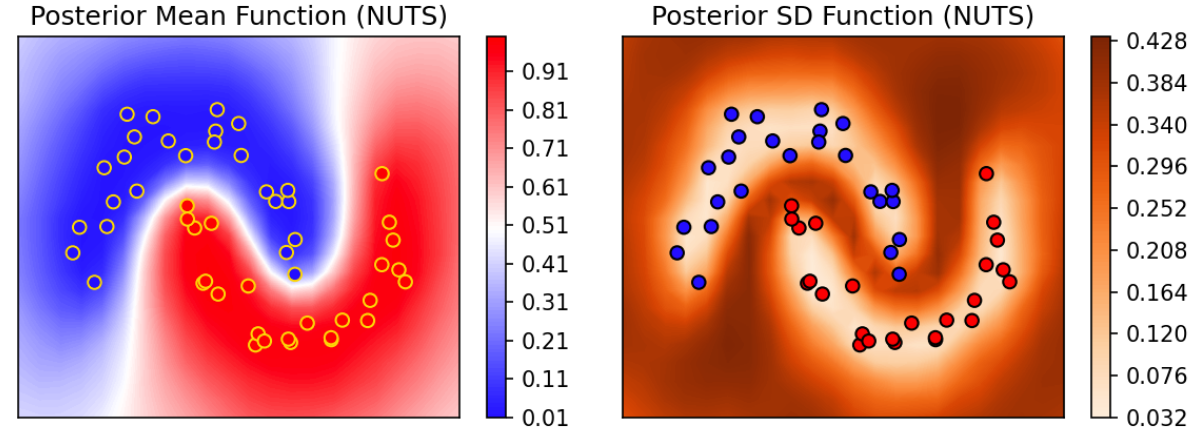
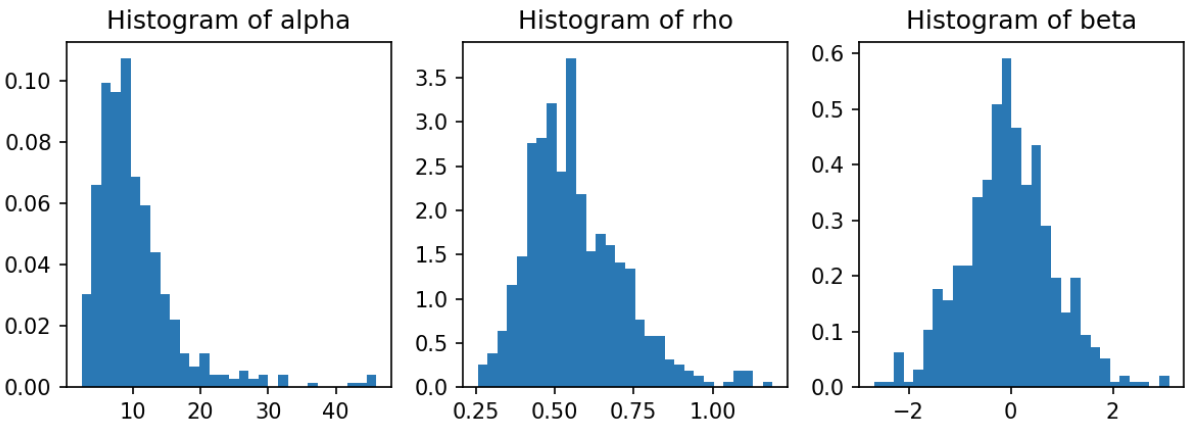
Note that one shortcoming of Turing, TFP, Pyro, and Numpyro is that the latent
function $f$ is not returned as posterior samples. Rather, due to the way the
model is specified, only $\eta$ is returned. This means that $f$ needs to be
recomputed. The re-computation of $f$ is not too onerous as the time spent
doing posterior prediction is dominated by the required matrix inversions (or
more efficient/stable variants using cholesky decompositions). Note that in
STAN, posterior samples of $f$ can be obtained using the transformed
parameters block.
Timings
Here we summarize timings for each aforementioned inference algorithm and PPL. Timings can be sorted by clicking the column-headers. The inference times for all algorithms are lowest in STAN. Turing has the highest inference times for all inference algorithms. All computations were done in a c5.xlarge AWS instance.
| PPL | ADVI (run) | HMC (run) | NUTS (run) | ADVI (compile) | HMC (compile) | NUTS (compile) |
|---|---|---|---|---|---|---|
| stan | 0.399 | 3.83 | 7.2 | 56.2 | 56.2 | 56.2 |
| turing | 13.176 | 64.383 | 102.772 | 15.272 | 20.159 | 11.685 |
| pyro | 5.0 | 31.0 | 73.0 | 0.0 | 0.0 | 0.0 |
| numpyro | 4.59 | 10.0 | 18.0 | 0.338 | 5.2 | 0.6 |
| tfp | 3.11 | 8.71 | 52.0 | 0.0 | 2.84 | 4.54 |